Amparo TolosaGenotyping
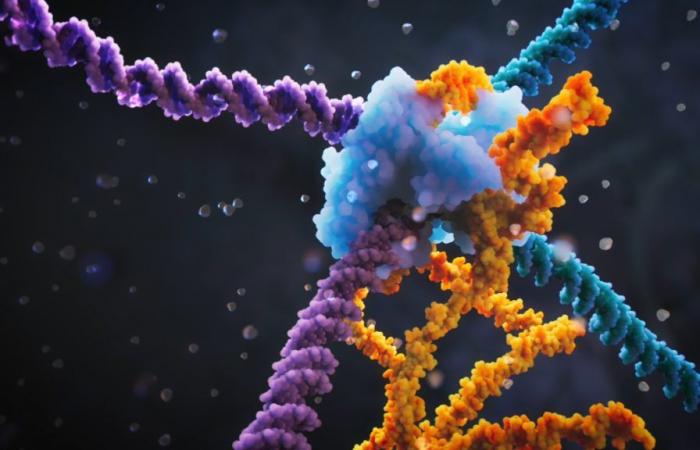
Researchers at the Arc Institute, the University of California Berkeley and the University of Tokyo have developed a new genome editing technique that allows the insertion, inversion or deletion of long DNA sequences at specific positions in the genome.
The recent genome editing tools such as CRISPR or base editors have opened a range of possibilities for biotechnology and medicine. These approaches are useful to carry out specific modifications in the genome efficiently. With them it is possible, for example, to change a DNA nucleotide to correct a mutation. This is precisely what the first approved genome editing therapy for sickle cell disease consists of. However, it is not possible to precisely make modifications that affect long fragments of the genome.
Two articles recently published in Nature offer a new genome editing solution for large fragments through a new technique based on a rProgrammable ecombinase directed by a small RNA molecule bridgeThis technology allows three basic types of DNA rearrangements to be carried out: insertions, excisions and inversions, at specific positions in the genome. The two studies demonstrate that this method is effective in bacteria and suggest its potential for other organisms and cells. If it works and is optimised, it could lead to treatments for diseases caused by chromosomal alterations.
“The bridging RNA system is a completely new mechanism for biological programming,” said Patrick Hsu, principal investigator at the Arc Institute and the University of California Berkeley and one of the directors of the work. “Bridging recombination can universally modify genetic material through insertion, excision, inversion and more, making possible a word processor for the living genome beyond CRISPR.”
Editing large DNA fragments from nature to the laboratory
The new system for modifying DNA uses a recombinase enzyme. These types of enzymes, present in nature, facilitate the cutting and joining of DNA sequences into new combinations. Therefore, they are very attractive for designing genome editing systems. Especially in the case of recombinases that recognize specific sequences in the DNA fragments involved.
In general, these recombinases recognize DNA sequences through a complex interaction system between protein and DNA. The novelty and main advantage of the system developed by researchers from the Arc Institute and the University of California Berkeley is that the recognition of specific DNA sequences involved in genetic rearrangement is mediated by a small bridging RNAwhich can be designed according to the needs of researchers.
The bridging RNA contains a region that specifies the source DNA sequence and another region that specifies the destination site in the DNA. And most importantly, both regions can be rescheduled. Specific RNAs can be created that recognize a wide range of DNA sequences and allow different types of reorganization of genetic material.
As occurred with the well-known CRISPR genome editing system, the new technique with RNA-guided recombinases derives from mobile genetic elements present in prokaryotic genomes, called IS110, which encode a recombinase and a bridging RNA.
Advantages of the new editing technique based on recombination with bridging RNA
The RNA bridge represents the first example of a bispecific molecule directed to a source DNA (DNA that is integrated) and a target DNA (where the DNA fragment is incorporated). Its combination with a recombinase to create a genome editing system offers multiple advantages.
The first of these is the already mentioned possibility of design the RNA for specific DNA sequences. Furthermore, it is a very compact systemwhich facilitates its use as an editing tool, since it can be easily packaged in vectors that transport it into cells. And finally, the system with the bridging RNA promotes the insertion, deletion or inversion of DNA fragments. in a clean wayin a clean way. The cuts and splices between the DNA strands involved are clean, without dragging part of the neighboring DNA. This is a significant improvement over less efficient long snippet editing methods.
“The bridging recombination mechanism solves some of the most fundamental challenges facing other genome editing methods,” said Matthew Durrant, a researcher at the Arc Institute in Palo Alto and the University of California Berkeley, as well as co-director of the investigation. “The ability to programmably rearrange any two DNA molecules opens the door to great advances in genome design.”
Great expectations to meet
At the moment, the editing system based on recombinases with bridging RNA has shown efficiency, although variable, in bacteria. Insertions have also been observed at undesirable points in the genome. It will therefore be necessary to optimize its functioning, and, above all, determine whether it also works in mammalian cells or more specifically, in humans.
“It is foreseeable that they will do so, although we should wait to see these results before ringing the bells too soon, for a recombination editing system based on IS mobile elements that promises to solve the shortcomings of CRISPR systems when it comes to reconstructing serious chromosomal alterations that are frequently the cause of congenital diseases,” said Lluís Montoliu, a researcher at the National Biotechnology Centre specialising in genome editing, who was not involved in the study. Science Media Center.
Also with the goal of developing tools for large-scale genome editing, researchers in David Liu’s lab at the Broad Institute of MIT, Harvard, and HHMI recently improved the gene editing technology called prime editing to insert or replace entire genes into human cells efficiently. Published in Nature Biomedical EngineeringThe eePASSIGE method combines prime editing with designer recombinase enzymes to insert long DNA sequences into specific sites in the genome.
Scientific articles:
Hiraizumi, M., Perry, N.T., Durrant, M.G. et al. Structural mechanism of bridge RNA-guided recombination. Nature 630994–1002. 2024. https://doi.org/10.1038/s41586-024-07570-2
Durrant, M.G., Perry, N.T., Pai, J.J. et al. Bridge RNAs direct programmable recombination of target and donor DNA. Nature 630, 984–993. 2024. https://doi.org/10.1038/s41586-024-07552-4
Pandey, S., Gao, X.D., Krasnow, N.A. et al. Efficient site-specific integration of large genes in mammalian cells via continuously evolved recombinases and prime editing. Nat. Biomed. Eng. 2024. https://doi.org/10.1038/s41551-024-01227-1
Other sources:
Arc Institute Scientists Discover Next-Generation System for Programmable Genome Design. https://arcinstitute.org/news/news/bridgeTou CJ, Kleinstiver BP. Programmable RNA-guided enzymes for next-generation genome editing. Nature. 2024 Jun;630(8018):827-828. doi: http://dx.doi.org/10.1038/d41586-024-01461-2